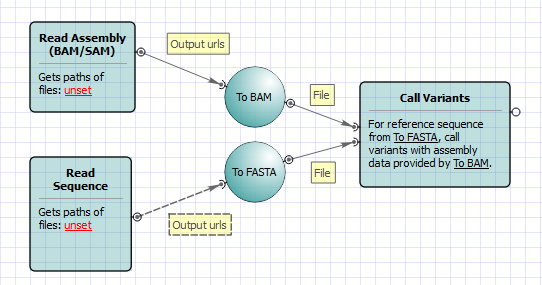
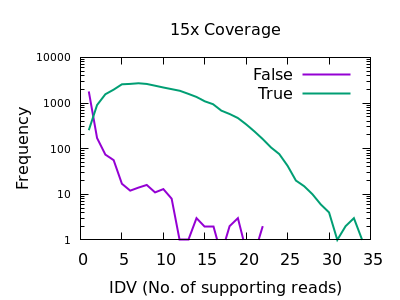
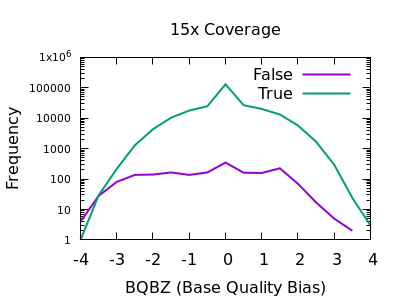
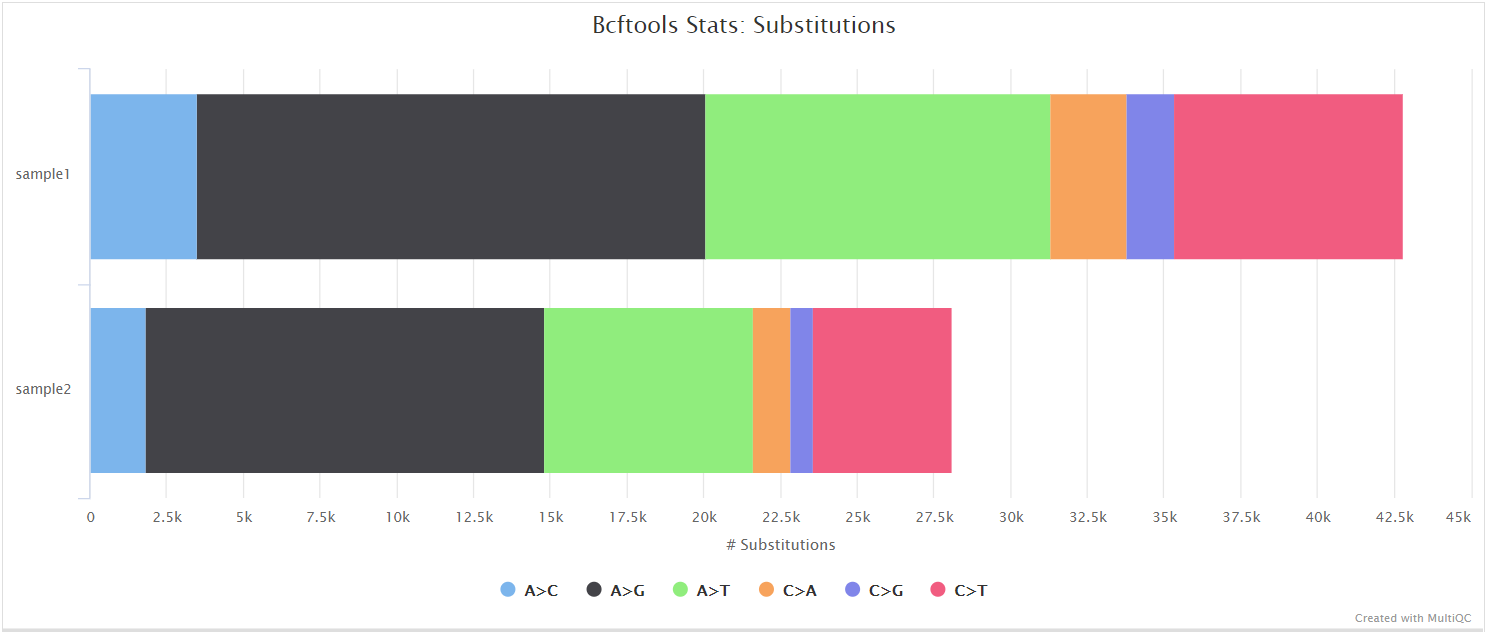
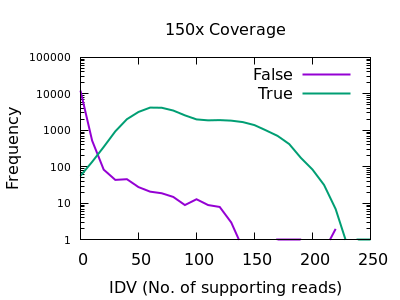
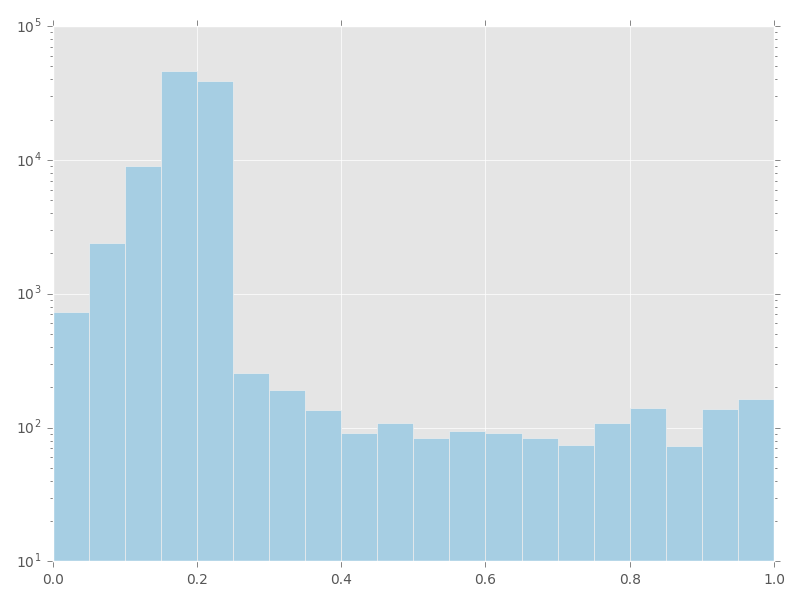
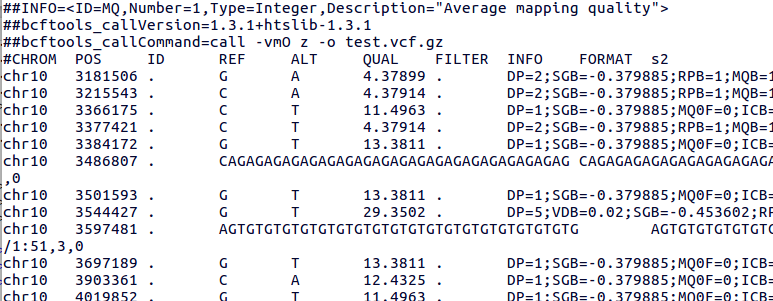
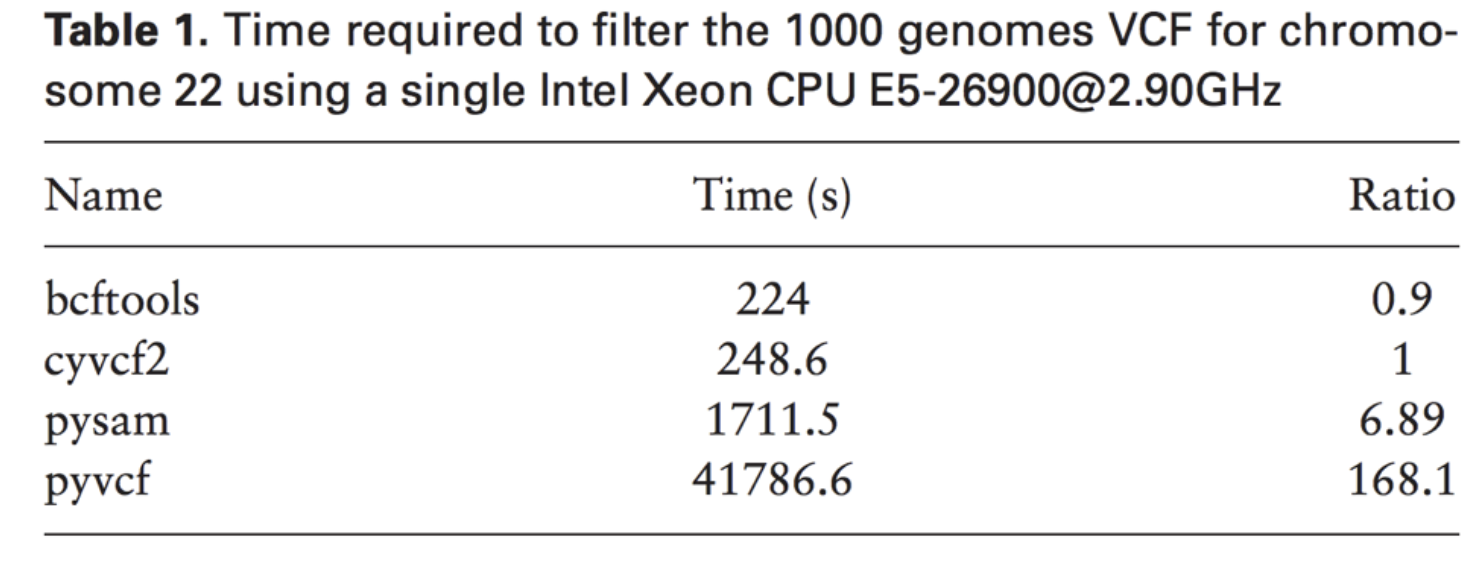
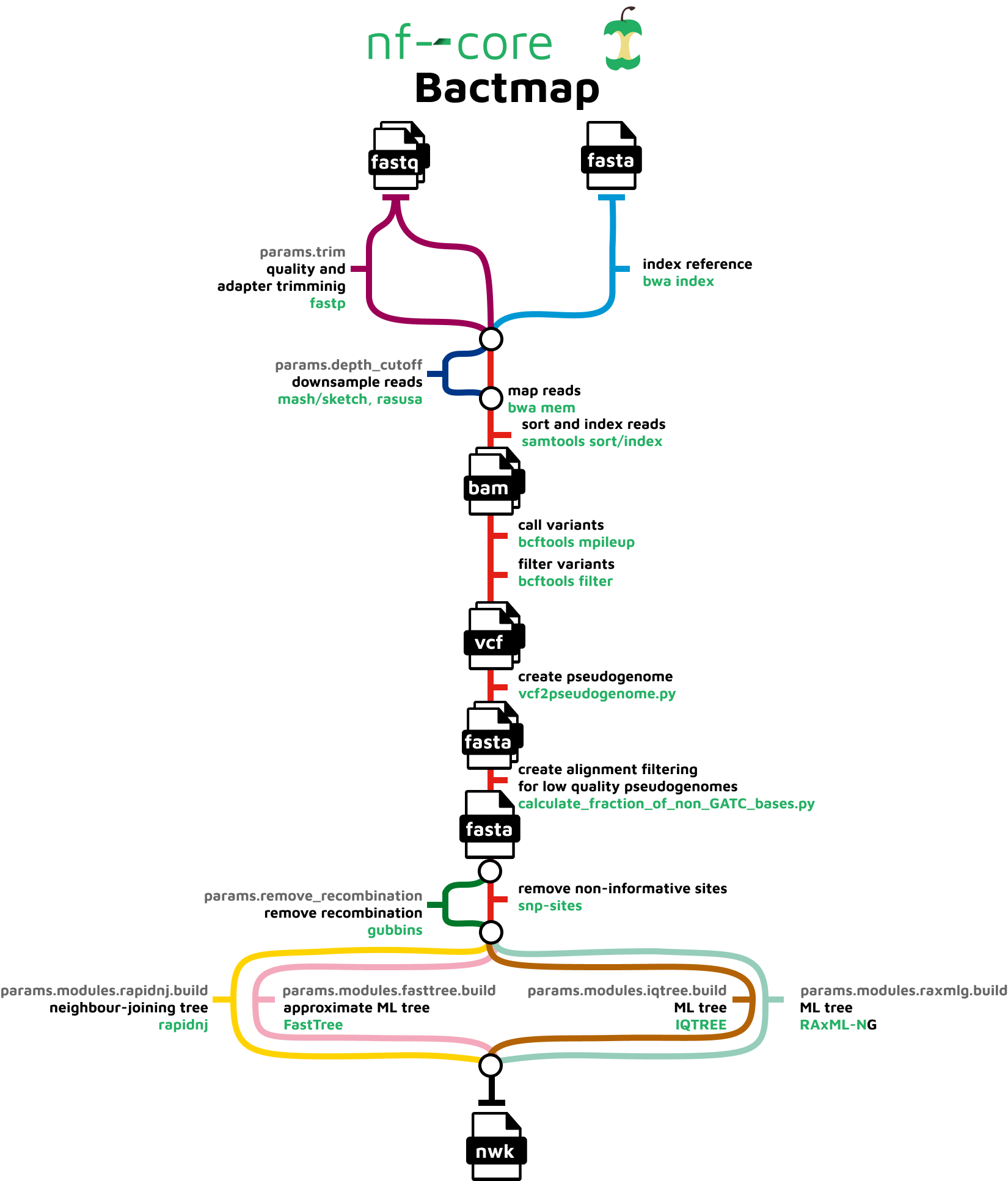
bcftools mpileup - Difference between IDV and FORMAT/AD* fields · Issue #912 · samtools/bcftools · GitHub

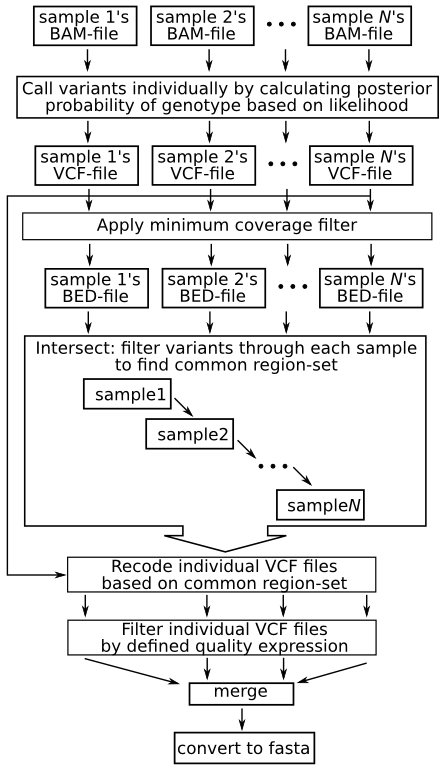
Variant calling using NGS and sequence capture data for population and evolutionary genomic inferences in Norway Spruce (Picea abies) | bioRxiv
Possible to annotate the FILTER from an annotation VCF file, as an INFO/TAG in the target? · Issue #1187 · samtools/bcftools · GitHub
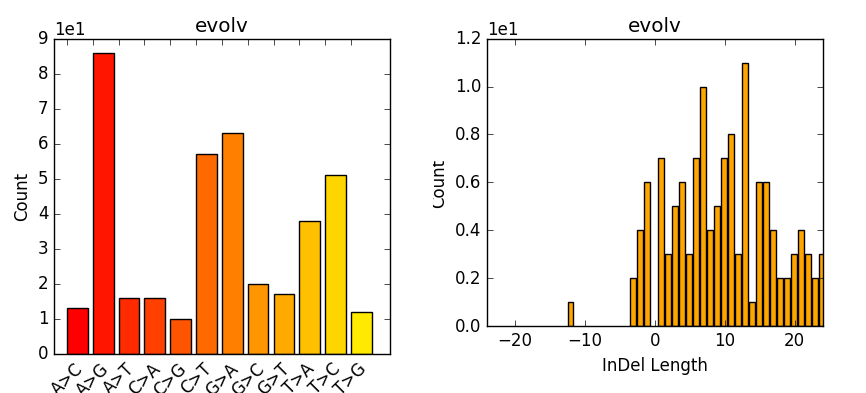
PLOS Genetics: No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice